Whole-brain PFC inputs (Le Merre and Ährlund-Richter, Neuron, 2021; methodology for Box 2)
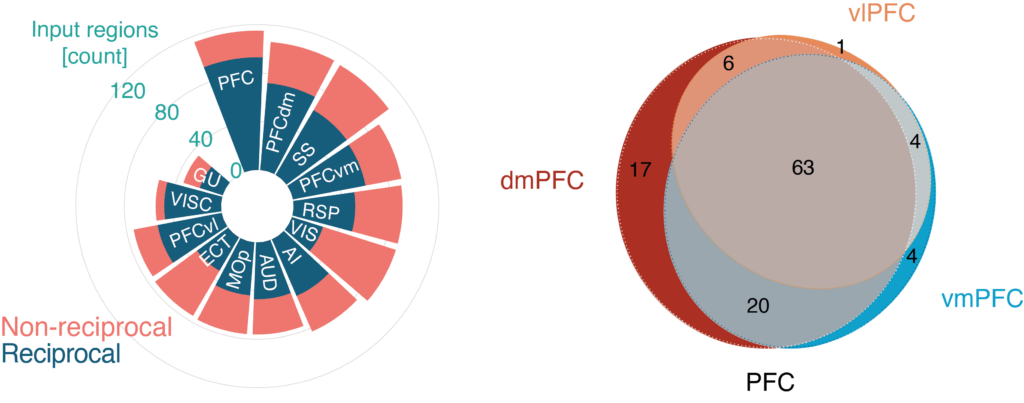
The circular plot displays the number of discrete brain regions giving input (coral red) to the whole mouse PFC, the three PFC subdivisions and different isocortical regions, respectively. Dark blue: the proportion of the projections being reciprocal. The regions are plotted from the largest to the smallest number of input regions. The Venn diagram displays the number of unique and shared input regions for the three PFC subdivisions. Gray; input regions (n = 63) shared by all three PFC subdivisions. Each experiment in the Allen Institute for Brain Science connectome with an injection volume below 0.5 mm3 and a confirmed projection volume of 0.02mm3 (volumes of voxels labeled as projecting) in a target area was counted as an input region. Reciprocal connectivity was confirmed by the identification of a projection volume of 0.02mm3 in the input region from the target region.
PFC Functional Research Synthesis (Le Merre and Ährlund-Richter, Neuron, 2021; methodology for figure 3)
Data and methodology used to perform the functional research synthesis presented in the figure 3 of the perspective article: The mouse prefrontal cortex: Unity in diversity. The code used to generate the figures can be found on this github repository
3D visualisation
Here is an attempt of 3D visualisation of the database using plotly. Some improvements are still required. The Task type and Sensory Modality parameters are displayed as continuous variables although they are discrete variables. You can mouse over the dots in the scatter plot to see from which study is the data ONLY if you have already scrolled within the brain mesh (plotly feature). We hope to improve this 3D visualisation with time.
Database
The database used to perform our research synthesis is available as a table. This table has the following columns: the year of the study, the short publication reference, the PFC region name used in the original publication, the inactivation method used, the stereotaxic triplet provided in the original article. The averaged coordinates used for the 3D plot into the Allen reference Atlas (ARA) CCFv3, the correction applied to the DV value to translate it into the ARA (see also below), the 3 scores; complexity index, sensory modality and task type, obtained for every publication, 7 logical values (in MOs, in ACA, in PL, in ILA, in ORBm, in ORBvl and in ORBl) indicating in which PFC region the study is found when plotted into the ARA CCFv3 with our method and the name of the Allen PFC region containing the stereotaxic triplet provided in the original publication.
The full database table can be downloaded here as an Excel or csv file.
| Year | Ref Short | Area Name | Inactivation Method | AP | ML | DV | DV origin | Mean AP | Mean ML | Mean DV | Corrected DV | Brain Surface DV Corr | Complexity index | Sensory Modality | Task type | in MOs | in ACA | inPL | in ILA | in ORBm | in ORBvl | in ORBl | Allen Atlas Annotation (2017) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2018 | Abbas et al., Neuron, 2018 | mPFC | Photoinhibition | 1,85 | 0,35 | 1,3 | DV brain surface | 1,85 | 0,35 | 1,3 | 2,065 | 0,765 | 4 | 6 | 3 | false | false | true | false | false | false | false | PL |
| 2017 | Allen et al., Neuron, 2017 | ALM | Photoinhibition, Muscimol | 2,4 | 2 | 0,5 | DV brain surface | 2,4 | 2 | 0,5 | 1,685 | 1,185 | 4 | 4 | 1 | true | false | false | false | false | false | false | MOs |
| 2018 | Allsop et al., Cell, 2018 | ACC | Phtoinhibition | 1 | 0,3 | 2,1 | DV bregma | 1 | 0,3 | 2,1 | 2,1 | 0 | 7 | 6 | 3 | false | true | false | false | false | false | false | ACA |
| 2018 | Baltz et al., 2018 | OFC | Photoinhibition (PV-ChR2)/ Chemogenetics (hMD4) | 2,7 | 1,67 | 2,65 | DV Bregma | 2,7 | 1,67 | 2,65 | 2,65 | 0 | 5 | 6 | 2 | true | false | false | false | false | false | false | ORBl |
| 2020 | Bari et al., 2020 | dmPFC | Photoactivation (ChR2) | 1,7 | 0,3 | 2 | DV brain surface | 1,7 | 0,3 | 2 | 2,745 | 0,745 | 7 | 2 | 3 | false | false | true | false | false | false | false | PL |
| 2019 | Bari et al., Neuron 2019 | mPFC | Muscimol (bilateral) | 2,5 | 0,5 | 1 | DV brain surface | 2,5 | 0,4 | 1,25 | 2,295 | 1,045 | 5 | 4 | 1 | false | false | true | false | false | false | false | PL |
| 2020 | Bariselli et al,. 2020 | OFC | Photoactivation (ChR2) | 2,5 | 1,1 | 2,5 | DV brain surface | 2,5 | 1,1 | 2,5 | 3,415 | 0,915 | 2 | 6 | 1 | false | false | false | false | false | false | true | ORBvl |
| 2017 | Barker et al., 2017 | IfL-C | Photoinhibition (arch) | 1,3-2 | 0,2-0,4 | 3 | DV Bregma | 1,65 | 0,3 | 3 | 3 | 0 | 3 | 6 | 2 | false | false | false | true | false | false | false | ILA |
| 2015 | Beier et al., Cell, 2015 | mPFC | Photoactivation | 1,95-2,25 | 0,27 | 1,6-2,1 | DV brain surface | 2,1 | 0,27 | 1,85 | 2,815 | 0,965 | 6 | 6 | 2 | false | false | false | true | false | false | false | ILA |
| 2019 | Berg et al., 2017 | IL | Photoactivation (Pyr- ChR2) | 1,66 | 0,3 | 1,6-2 | DV brain surface | 1,66 | 0,3 | 1,8 | 2,535 | 0,735 | 2 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2017 | Bolkan et al., Nat Neurosci, 2017 | mPFC | Phtoinhibition | 1,75 | 0,4 | 1,8 | DV brain surface | 1,75 | 0,4 | 1,8 | 2,485 | 0,685 | 5 | 6 | 3 | false | false | true | false | false | false | false | PL |
| 2019 | Breton-Provencher and Sur, Nat Neurosci, 2019 | OFC | Photoactivation | 2,6 | 1,3 | 2 | DV bregma | 2,6 | 1,3 | 2 | 2 | 0 | 1 | 1 | 1 | false | false | false | false | false | false | true | ORBl |
| 2015 | Bukalo et al., Neuropsychology, 2015 | vmPFC | Photoinhibition | 1,8 | 0,3 | 2,8 | DV bregma | 1,8 | 0,3 | 2,8 | 2,8 | 0 | 5 | 6 | 2 | false | false | false | true | false | false | false | ILA |
| 2017 | Carus-Cadavieco et al., 2017 | mPFC | Photostimulation and inhibition (eNPAC2) | 1,7 | 0,35 | 2,7 | DV Bregma | 1,7 | 0,35 | 2,7 | 2,7 | 0 | 2 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2019 | Cho et al., BioRxiv, 2019 | mPFC | Photoactivation | 1,7 | 0,3 | 2,75 | DV bregma | 1,7 | 0,3 | 2,75 | 2,75 | 0 | 4 | 6 | 2 | false | false | true | true | false | false | false | ILA |
| 2015 | Cho et al., Neuron, 2015 | mPFC | Photoinhibition (arch) | 1,7 | 0,3 | 2,75 | DV bregma | 1,7 | 0,3 | 2,75 | 2,75 | 0 | 4 | 6 | 2 | false | false | true | true | false | false | false | ILA |
| 2014 | Courtin et al., Nature, 2014 | dmPFC | Photoinhibition | 2 | 0,3 | 0,8-1,4 | DV brain surface | 2 | 0,3 | 1,1 | 1,985 | 0,885 | 6 | 1 | 2 | false | false | true | false | false | false | false | PL |
| 2016 | Dejean et al., 2016 | dmPFC | Photoinhibition (PV-ChR2) | 2 | 0,3 | 0,8-1,4 | DV brain surface | 2 | 0,3 | 1,2 | 2,085 | 0,885 | 5 | 1 | 3 | false | false | true | false | false | false | false | PL |
| 2020 | Esmaeili et al., 2020 | mPFC | Photoinhibition | 2 | 0,5 | 1,7 | DV brain surface | 2 | 0,5 | 1,7 | 2,435 | 0,735 | 5 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2016 | Felix-Ortiz et al., Neuroscience, 2016 | mPFC | Photostimulation | 1,7 | 0,3 | 1,9 | DV bregma | 1,7 | 0,3 | 1,9 | 1,9 | 0 | 6 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2018 | Gilad et al., Neuron, 2018 | M2 | Photoinhibition | 1,5 | 0,5 | 0,5 | DV brain surface | 1,5 | 0,5 | 0,5 | 1,035 | 0,535 | 6 | 3 | 1 | true | false | false | false | false | false | false | MOs |
| 2016 | Goard et al., Elife, 2016 | fMC | Photoinhibition | 1,5 | 1 | 0,5 | DV brain surface | 1,5 | 1 | 0,5 | 0,995 | 0,495 | 6 | 2 | 1 | true | false | false | false | false | false | false | MOs |
| 2012 | Granon and Changeux, Behav Brain Res, 2012 | PL | NMDA lesion | 1,9-2,8 | 0,3-0,4 | 1,5-2,5 | DV brain surface | 2,35 | 0,35 | 2 | 3,005 | 1,005 | 4 | 6 | 3 | false | false | false | false | true | false | false | ORBm |
| 2014 | Guo et al., Neuron, 2014 | ALM | Photoinhibition | 2,5 | 1,5 | 0,5 | DV brain surface | 2,5 | 1,5 | 0,5 | 1,525 | 1,025 | 7 | 3 | 1 | true | false | false | false | false | false | false | MOs |
| 2020 | Gutzeit et al., 2020 | IL-PFC | Photoactivation (ChR2) | 1,8 | 0,3 | 2,3 | DV Bregma | 1,8 | 0,3 | 2,3 | 2,3 | 0 | 5 | 6 | 3 | false | false | true | false | false | false | false | PL |
| 2020 | Halladay et al., 2020 | vmPFC | Photoinhibition (Arch) | 1,5-2,5 | 0,5 | 2,7 | DV Bregma | 2 | 0,5 | 2,7 | 2,7 | 0 | 5 | 6 | 3 | false | false | true | true | false | false | false | ILA |
| 2019 | Hare et al., 2019 | PL - IL | Photostimulation - DREADD (hM4D) | 1,9 | 0,4 | 2,5 | DV Bregma | 1,9 | 0,4 | 2,5 | 2,5 | 0 | 3 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2020 | Heikenfeld et al., 2020 | mPFC | Photostimulation and inhibition (eNPAC2) | 1,5 | 0,3 | 3,1 | DV Bregma | 1,5 | 0,3 | 3,1 | 3,1 | 0 | 4 | 6 | 3 | false | false | false | true | false | false | false | ILA |
| 2019 | Hu et al., Neuron, 2019 | Cg | Photoinhibition, Photoactivation | 0,3 | 0,3 | 1 | DV brain surface | 0,3 | 0,3 | 1 | 1,235 | 0,235 | 5 | 2 | 1 | false | true | false | false | false | false | false | ACA |
| 2019 | Huang et al., 2019 | PL | Photoinhibition (NpHR) - Pharmacological ablation (dtA) | 1,9 | 0,5 | 2,1 | DV brain surface | 1,9 | 0,5 | 2,1 | 2,795 | 0,695 | 3 | 3 | 1 | false | false | true | true | false | false | false | ILA |
| 2018 | Huda et al., BioRxiv, 2018 | ACC | Photoinhibition | 0,5 | 0,5 | 0,5 | DV brain surface | 0,5 | 0,5 | 0,5 | 0,705 | 0,205 | 8 | 2 | 1 | true | false | false | false | false | false | false | MOs |
| 2018 | Inagaki et al., J.Neurosci, 2018 | ALM | Photoinhibition | 2,5 | 1,5 | 0,5 | DV brain surface | 2,5 | 1,5 | 0,5 | 1,525 | 1,025 | 6 | 1 | 1 | true | false | false | false | false | false | false | MOs |
| 2019 | Inagaki et al., Nature, 2019 | ALM | Photoinhibition | 2,5 | 1,5 | 0,5 | DV brain surface | 2,5 | 1,5 | 0,5 | 1,525 | 1,025 | 6 | 1 | 1 | true | false | false | false | false | false | false | MOs |
| 2018 | Itokazu et al., Nat Commun, 2018 | MOs | Photoinhibition | 0,7 | 0,7 | 0,25 | DV brain surface | 0,7 | 0,7 | 0,25 | 0,495 | 0,245 | 6 | 2 | 1 | true | false | false | false | false | false | false | MOs |
| 2019 | Jennings et al., Nature, 2019 | OFC | Photoactivation | 2,6 | 1,26 | 2,8 | DV bregma | 2,6 | 1,26 | 2,8 | 2,8 | 0 | 4 | 6 | 1 | false | false | false | false | false | false | true | ORBl |
| 2018 | Jhang et al., 2018 | ACC | Photoactivation and inhibition (eNpHR3, ChR2) | 0,8 | 0,35 | 1,5 | DV Bregma | 0,8 | 0,35 | 1,5 | 1,5 | 0 | 3 | 4 | 2 | false | true | false | false | false | false | false | ACA |
| 2017 | Kamigaki and Dan, Nat Neurosci, 2017 | dmPFC | Photoinhibition | 1,8-2,1 | 0,3-0,4 | 0,8-1,2 | DV brain surface | 1,95 | 0,35 | 1 | 1,805 | 0,805 | 4 | 1 | 1 | false | true | false | false | false | false | false | ACA |
| 2016 | Kim et al., 2016 | IL | Photoactivation and inhibition (eNpHR3, ChR2) | 1,65 | 0,3 | 3,05 | DV Bregma | 1,65 | 0,3 | 3,05 | 3,05 | 0 | 5 | 1 | 3 | false | false | false | true | false | false | false | ILA |
| 2016 | Kim et al., Cell, 2016a | mPFC | Photoactivation | 1,9 | 0,4 | 1 | DV brain surface | 1,9 | 0,4 | 1 | 1,755 | 0,755 | 8 | 6 | 3 | false | true | false | false | false | false | false | ACA |
| 2016 | Kim et al., Cell, 2016b | mPFC | Photoactivation | 1,76 | 0,25 | 1,25 | DV brain surface | 1,76 | 0,25 | 1,25 | 2,075 | 0,825 | 8 | 2 | 3 | false | false | true | false | false | false | false | PL |
| 2017 | Kim et al., Cell, 2017 | mPFC | Photoinhibition | 2,1 | 0,35 | 2,2-2,3 | DV bregma | 2,1 | 0,35 | 2,25 | 2,25 | 0 | 4 | 1 | 1 | false | false | true | false | false | false | false | PL |
| 2020 | Kingsbury et al., 2020 | dmPFC | Photoactivation (ChR2) | 2 | 0,3 | 1,8 | DV brain surface | 2 | 0,3 | 1,8 | 2,685 | 0,885 | 5 | 6 | 1 | false | false | true | true | false | false | false | ILA |
| 2017 | Klavir et al., Nat Neurosci, 2017 | mPFC | Photosactivation | 2 | 0,25 | 2,25 | DV bregma | 2 | 0,25 | 2,25 | 2,25 | 0 | 5 | 1 | 2 | false | false | true | false | false | false | false | PL |
| 2016 | Koike et al., Neuropsychopharmacology, 2016 | dACC | DREADD (hM4Di) | 1,7-1,1-0,4 | 0,2-0,2-0,4 | 0,7 | DV brain surface | 1,05 | 0,3 | 0,7 | 1,195 | 0,495 | 10 | 2 | 3 | false | true | false | false | false | false | false | ACA |
| 2013 | Kumar et al., 2013 | PrL | Photoactivation (ChR2, thy1-cre) | 2,65 | 0,25 | 0,8 | DV brain surface | 2,65 | 0,25 | 0,8 | 2,085 | 1,285 | 2 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2018 | Lak et al., BioRxiv, 2018 | PL | VTA photoinhibition | 1,8 | 0,3 | 1,4 | DV brain surface | 1,8 | 0,3 | 1,4 | 2,185 | 0,785 | 7 | 2 | 1 | false | false | true | false | false | false | false | PL |
| 2018 | Le Merre et al., Neuron, 2018 | mPFC | Photoinhibition, Muscimol | 1,8 | 0,3 | 1,75 | DV brain surface | 1,8 | 0,3 | 1,75 | 2,535 | 0,785 | 2 | 3 | 1 | false | false | true | false | false | false | false | PL |
| 2019 | Lee et al., Neuron, 2019 | mPFC | Photoinhibition (arch) | 1,7 | 0,3 | 2,75 | DV bregma | 1,7 | 0,3 | 2,75 | 2,75 | 0 | 2 | 6 | 1 | false | false | true | true | false | false | false | ILA |
| 2017 | Leinweber et al., Neuron, 2017 | A24b | Muscimol | 1 | 0,3 | 0,5 | DV brain surface | 1 | 0,3 | 0,5 | 0,975 | 0,475 | 6 | 2 | 1 | true | false | false | false | false | false | false | MOs |
| 2020 | Li et al., 2020 | dmPFC | Photoactivation and inhibition (iC++, ChR2) | 1 | 0,5 | 0,8 | DV brain surface | 1 | 0,5 | 0,8 | 1,165 | 0,365 | 5 | 1 | 1 | true | false | false | false | false | false | false | MOs |
| 2015 | Li et al., Nature, 2015 | ALM | Photoinhibition | 2,5 | 1,5 | 0,5 | DV brain surface | 2,5 | 1,5 | 0,5 | 1,525 | 1,025 | 7 | 3 | 1 | true | false | false | false | false | false | false | MOs |
| 2016 | Li et al., Nature, 2016 | ALM | Photoinhibition | 2,5 | 1,5 | 0,5 | DV brain surface | 2,5 | 1,5 | 0,5 | 1,525 | 1,025 | 7 | 3 | 1 | true | false | false | false | false | false | false | MOs |
| 2016 | Liu et al., 2016 | IL | SSFO | 1,75 | 0,45 | 2,8 | DV Bregma | 1,75 | 0,45 | 2,8 | 2,8 | 0 | 4 | 6 | 2 | false | false | true | true | false | false | false | ILA |
| 2014 | Liu et al., Science, 2014 | mPFC | Photoinhibition, DREADD (hM4Di) | 1,96 | 0,42 | 1,62 | DV brain surface | 1,96 | 0,42 | 1,62 | 2,385 | 0,765 | 6 | 4 | 3 | false | false | true | false | false | false | false | PL |
| 2017 | Makino et al., Neuron, 2017 | M2 | Muscimol | 2,6 | 1 | 0,5 | DV brain surface | 2,6 | 1 | 0,5 | 1,455 | 0,955 | 4 | 1 | 1 | true | false | false | false | false | false | false | MOs |
| 2015 | Manita et al., Neuron, 2015 | M2 | Photoinhibition, TTX, CNQX | 2,19 | 0,63 | 0,5 | DV brain surface | 2,19 | 0,63 | 0,5 | 1,275 | 0,775 | 3 | 3 | 1 | true | false | false | false | false | false | false | MOs |
| 2015 | Manita et al., Neuron, 2015 | M2 | Photoinhibition, TTX, CNQX | 2,19 | 0,63 | 0,5 | DV brain surface | 2,19 | 0,63 | 0,5 | 1,275 | 0,775 | 5 | 3 | 1 | true | false | false | false | false | false | false | MOs |
| 2018 | Marton et al., J. of Neuro, 2018 | mPFC | Photoinhibition | 1,7 | 0,3 | 2,75 | DV bregma | 1,7 | 0,3 | 2,75 | 2,75 | 0 | 4 | 6 | 2 | false | false | true | true | false | false | false | ILA |
| 2019 | Mayrhofer et al., Neuron, 2019 | ALM | Photoinhibition | 2,5 | 1,5 | 0,5 | DV brain surface | 2,5 | 1,5 | 0,5 | 1,525 | 1,025 | 3 | 6 | 1 | true | false | false | false | false | false | false | MOs |
| 2017 | Morandel et al., Scientific reports, 2017 | RFA | Photoinhibition | 2,7 | 0,9 | 0,5 | DV brain surface | 2,7 | 0,9 | 0,5 | 1,505 | 1,005 | 9 | 3 | 1 | true | false | false | false | false | false | false | MOs |
| 2012 | Morawska and Fendt, 2012 | mPFC | Muscimol | 1,6 | 0,5 | 2,1 | DV brain surface | 1,6 | 0,5 | 2,1 | 2,675 | 0,575 | 5 | 1 | 3 | false | false | true | false | false | false | false | PL |
| 2017 | Murugan et al., Cell, 2017 | PL | Photoinhibition | 1,8 | 0,5 | 2,5 | DV bregma | 1,8 | 0,5 | 2,5 | 2,5 | 0 | 6 | 6 | 3 | false | false | true | false | false | false | false | PL |
| 2019 | Nakajima et al., 2019 | PFC | Photoinhibition (eArch3) | 2,6 | 0,6 | 1 | DV brain surface | 2,6 | 0,6 | 1 | 1,995 | 0,995 | 10 | 6 | 2 | false | false | true | false | false | false | false | PL |
| 2018 | Nakayama et al., 2018 | mPFC | Photoactivation and inhibition (Arch, ChR2) | 1,9-2,0 | 0,5 | 1,6 | DV brain surface | 1,95 | 0,5 | 1,6 | 2,315 | 0,715 | 4 | 6 | 2 | false | false | true | false | false | false | false | PL |
| 2019 | Namboodiri et al., 2019 | vmOFC | Photoinhibition (NpHR3) | 2,5-2,9 | 1-1,1 | 2,3 | DV brain surface | 2,7 | 1,05 | 2,3 | 3,315 | 1,015 | 4 | 1 | 1 | false | false | false | false | false | true | true | ORBvl |
| 2019 | Newmyer et al., 2019 | IL | Photoactivation (ChR2, SSFO) | 1,8 | 0,4 | 1,8 | DV brain surface | 1,8 | 0,4 | 1,8 | 2,505 | 0,705 | 2 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2020 | Nguyen et al., 2020 | mPFC | Photoactivation and inhibition (Arch, ChR2) | 1,9 | 0,3 | 2,3 | DV Bregma | 1,9 | 0,3 | 2,3 | 2,3 | 0 | 5 | 4 | 3 | false | false | true | false | false | false | false | PL |
| 2017 | Otis et al., Nature, 2017 | PFC | Photoinhibition, Photoactivation | 1,85 | 0,6 | 2,5 | DV bregma | 1,85 | 0,6 | 2,5 | 2,5 | 0 | 4 | 1 | 1 | false | false | true | false | false | false | false | PL |
| 2019 | Padilla-Coreano et al., Neuron, 2019 | mPFC | Photoactivation | 1,6 | 0,4 | 1,25 | DV brain surface | 1,6 | 0,4 | 1,25 | 1,885 | 0,635 | 2 | 6 | 1 | false | true | true | false | false | false | false | PL |
| 2018 | Pascoli et al., Nature, 2018 | OFC | Photoactivation | 2,6 | 1,6 | 1,8 | DV brain surface | 2,6 | 1,6 | 1,8 | 2,905 | 1,105 | 5 | 6 | 2 | false | false | false | false | false | false | true | ORBl |
| 2015 | Pinto and Dan, Neuron, 2015 | dmPFC | Muscimol | 2,1 | 0,3 | 1,6 | DV brain surface | 2,1 | 0,3 | 1,6 | 2,535 | 0,935 | 5 | 1 | 1 | false | false | true | false | false | false | false | PL |
| 2019 | Pinto et al., Cell, 2019 | aM2 | Photoinhibition | 2,5 | 0,25 | 0,5 | DV brain surface | 2,5 | 0,25 | 0,5 | 1,705 | 1,205 | 6 | 2 | 1 | false | true | false | false | false | false | false | ACA |
| 2019 | Pinto et al., Cell, 2019 | aM2 | Photoinhibition | 2,5 | 0,25 | 0,5 | DV brain surface | 2,5 | 0,25 | 0,5 | 1,705 | 1,205 | 7 | 2 | 1 | false | true | false | false | false | false | false | ACA |
| 2016 | Popescu et al., PNAS, 2016 | mPFC | Photoactivation (DA) | 1,97 | 0,4 | 1,6-1,7 | DV brain surface | 1,97 | 0,4 | 1,65 | 2,435 | 0,785 | 2 | 1 | 1 | false | false | true | false | false | false | false | PL |
| 2015 | Rajasethupathy et al., 2015 | AC | Photoactivation and inhibition (eNpHR3, ChR2) | 1 | 0,3 | 1,2 | DV brain surface | 1 | 0,3 | 1,2 | 1,675 | 0,475 | 6 | 6 | 3 | false | true | false | false | false | false | false | ACA |
| 2018 | Rikhye et al., Nat Neurosci, 2018 | PFC | Photoinhibition | 2,1-2,7 | 0,25-0,6 | 1,2-1,7 (ant) and 2-2,4 (post) | DV brain surface | 2,4 | 0,43 | 1,83 | 2,805 | 0,975 | 10 | 6 | 2 | false | false | false | false | true | false | false | ORBm |
| 2018 | Rozeske et al., Neuron, 2018 | dmPFC | Photoinhibition | 1,98 | 0,35 | 1,5 | DV brain surface | 1,98 | 0,35 | 1,5 | 2,325 | 0,825 | 7 | 6 | 2 | false | false | true | false | false | false | false | PL |
| 2017 | Schmitt et al., Nature, 2017 | PFC | Photoinhibition, SSFO | 2 | 0,6 | 1,2-1,7 (ant) and 2-2,4 (post) | DV brain surface | 2 | 0,6 | 1,83 | 2,525 | 0,695 | 10 | 6 | 2 | false | false | true | false | false | false | false | PL |
| 2014 | Schneider et al., Nature, 2014 | M2 | Photoactivation | 1 | 0,5 | 0,5 | DV brain surface | 1 | 0,5 | 0,5 | 0,865 | 0,365 | 2 | 6 | 1 | true | false | false | false | false | false | false | MOs |
| 2020 | Sheggia et al., 2020 | mPFC | Photoactivation and inhibition (eNpHR3, ChR2) | 1,9 | 0,3 | 2,4 | DV Bregma | 1,9 | 0,3 | 2,4 | 2,4 | 0 | 5 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2016 | Siniscalchi et al, Nat Neurosci, 2016 | M2 | Muscimol | 1,5 | 0,5 | 0.5 | DV brain surface | 1,5 | 0,5 | 0,5 | 1,035 | 0,535 | 7 | 1 | 2 | true | false | false | false | false | false | false | MOs |
| 2019 | Spellman et al., BioRxiv, 2019 | PFC | Muscimol / Photoinhibition (stGtACR2) | 1,75 | 0,35 | 1,75 | DV brain surface | 1,75 | 0,35 | 1,75 | 2,475 | 0,725 | 7 | 6 | 2 | false | false | true | false | false | false | false | PL |
| 2015 | Spellman et al., Nature, 2015 | mPFC | Photoinhibition | 1,8 | 0,4 | 1,4 | DV brain surface | 1,8 | 0,4 | 1,4 | 2,105 | 0,705 | 4 | 1 | 3 | false | false | true | false | false | false | false | PL |
| 2018 | Starkweather et al., Neuron, 2018 | mPFC | DREADD (KORD) | 2,7 | 0,25 | 0,6 | DV brain surface | 2,7 | 0,25 | 0,6 | 1,925 | 1,325 | 3 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2017 | Tamura et al., Nat Commun, 2017 | mPFC | Photoinhibition | 1,6 | 0,3 | 1,4 | DV brain surface | 1,6 | 0,3 | 1,4 | 2,105 | 0,705 | 4 | 4 | 3 | false | false | true | false | false | false | false | PL |
| 2019 | Tan et al., 2019 | PFC | Photoactivation (ChR2) | 2,25 | 0,45 | 1,2 | DV brain surface | 2,25 | 0,45 | 1,2 | 2,075 | 0,875 | 6 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2018 | Tuscher et al., 2018 | mPFC | DREADD (hM4Di) | 1,8 | 0,3 | 2,7 | DV Bregma | 1,8 | 0,3 | 2,7 | 2,7 | 0 | 5 | 6 | 1 | false | false | true | false | false | false | false | ILA |
| 2013 | Van den Oever et al., 2013 | vmPFC | Photoactivation and inhibition (eNpHR3, ChR2) | 2,2 | 0,4 | 3 | DV Bregma | 2,2 | 0,4 | 3 | 3 | 0 | 5 | 6 | 3 | false | false | false | false | true | false | false | ORBm |
| 2020 | Vertechi et al., 2020 | OFC | Photoinactivation (ChR2, Vgat) | 2,9 | 1,25 | 1,8 | DV brain surface | 2,9 | 1,25 | 1,8 | 2,965 | 1,165 | 3 | 6 | 1 | false | false | false | false | false | false | true | ORBl |
| 2018 | White et al., Cell Rep, 2018 | ACC | Photoinhibition | 0,74-1,34 | 0,3 | 1-1,25 | DV bregma | 1,04 | 0,3 | 1,13 | 1,13 | 0 | 10 | 6 | 3 | true | false | false | false | false | false | false | MOs |
| 2015 | Wimmer et al., Nature, 2015 | mPFC | Photoinhibition | 2,6 | 0,25 | 1,25 | DV brain surface | 2,6 | 0,25 | 1,25 | 2,505 | 1,255 | 10 | 1 | 2 | false | false | true | false | true | false | false | PL |
| 2020 | Wu et al., 2020 | ALM | Photoinhibition | 2,5 | 1,5 | 1,1 | DV Bregma | 2,5 | 1,5 | 1,1 | 1,1 | 0 | 8 | 4 | 3 | true | false | false | false | false | false | false | MOs |
| 2019 | Xu et al., BioRxiv 2019 | ALM | Photoinhibition | 2,2 | 1,5 | 1,3 | DV brain surface | 2,2 | 1,5 | 1,3 | 2,185 | 0,885 | 3 | 3 | 1 | true | false | false | false | false | false | false | MOs |
| 2020 | Yamamuro et al., 2020 | mPFC | Photoactivation and inhibition (eNpHR3, ChR2), DREADD )hMD4) | 1,3-2,3 | 0,4 | 1-1,7 | DV brain surface | 1,8 | 0,4 | 1,35 | 2,055 | 0,705 | 6 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2016 | Ye et al., Cell, 2016 | mPFC | Photoactivation | 1,9 | 0,35 | 2,6 | DV bregma | 1,9 | 0,35 | 2,6 | 2,6 | 0 | 2 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2011 | Yizhar et al., Nature, 2011 | mPFC | SSFO | 1,8 | 0,35 | 2,0-2,85 | DV bregma | 1,8 | 0,35 | 2,43 | 2,43 | 0 | 5 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2015 | Zhang et al., 2015 | PrL | Photoactivation and inhibition (Arch, ChR2) | 1,8 | 0,5 | 2,2 | DV Bregma | 1,8 | 0,5 | 2,2 | 2,2 | 0 | 4 | 6 | 1 | false | false | true | false | false | false | false | PL |
| 2014 | Zhang et al., Science, 2014 | Cg | Photoactivation | 0,2 | 0,3 | 0,9 | DV brain surface | 0,2 | 0,3 | 0,9 | 1,105 | 0,205 | 5 | 1 | 1 | false | true | false | false | false | false | false | ACA |
| 2020 | Zhu et al., 2020 | PrL | Photoinhibition | 1,98 | 0,4 | 1,65 | DV brain surface | 1,98 | 0,4 | 1,65 | 2,435 | 0,785 | 6 | 4 | 3 | false | false | true | false | false | false | false | PL |
| 2017 | Zimmermann et al., Biol. Psychiatry, 2017 | OFC | DREADD (BDNF KO) | 2,5 | 1,7 | 1,8 | DV bregma | 2,5 | 1,7 | 1,8 | 1,8 | 0 | 4 | 6 | 2 | true | false | false | false | false | false | false | MOs |
Details on the sensory modality, task type and complexity index
The primary sensory modality utilized in the studies was simply reported as a score from 1 to 6; auditory (1), visual (2), somatosensory (3), olfactory (4), gustatory (5) or multisensory (combination of two or more modalities; 6), respectively. The task type in the studies was also reported with a score from 1 to 3; sensorimotor transformation (1), context/rule (2), or memory/delay (3), respectively. A complexity index (ranging from 1 to 10) was calculated for each study. The behavioral task in each study was scored according to eight criteria: number of stimuli, complexity of the stimuli (0, 1, or 2), number of actions to perform, complexity of the action to perform (0, 1, or 2), presence of a cue indicating the beginning of a trial (0 or 1), presence of a delay between task events (0 or 1), presence of a short- or long-term memory component (0 or 1), freely moving (1) or head-fixed (0) task.
The full table can be downloaded here as an Excel or csv file.
| Ref Short | Task | Number of Stimuli | Complexity of the Stimuli | Number of Actions | Complexity of the Action | Cue | Delay | Memory Component (long term) | Freely Moving /HeadFixed | FINAL SCORE |
|---|---|---|---|---|---|---|---|---|---|---|
| Abbas et al., Neuron, 2018 | spatial Working memory : delayed non-match-to-place task. | 0 (no) | 0 (no) | 2 (go right or left) | 0 (normal behavior) | 0 (no) | 1(10-60s) | 0 (no) | 1 | 4 |
| Allen et al., Neuron, 2017 | olfactory go/no-go decision-making task | 2 (odors) | 0 (pure odor) | 1 (licking) | 0 (normal behavior) | 1 (Sound) | 0 (no) | 0 (no) | 0 | 4 |
| Allsop et al., Cell, 2018 | spatial Working memory : delayed non-match-to-place task. | 2 mouse being shocked in a context (fear cage) | 0 (pure) | 1 (freezing) | 0 (normal behavior) | 2 (compound light and 10kHz tone) | 0 (no) | 1 (24h) | 1 | 7 |
| Baltz et al., 2018 | incentive learning task | 0 (no) | 0 (no) | 3 (2 lever press right-left, licking central port) | 1 (motor sequence) | 0 (no) | 0 (no) | 0 (no) | 1 | 5 |
| Bari et al., 2020 | 2-choice task | 2 (lights) | 0 (flashing LED) | 2 (go right or left) | 0 (normal behavior) | 1 (reward port light at trial onset) | 1 (5s) | 0 (no) | 1 | 7 |
| Bari et al., Neuron 2019 | dynamic foraging | 2 (odors) | 0 (pure odor) | 2 (lick right or left) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (previous trial) | 0 | 5 |
| Bariselli et al,. 2020 | Open field on cocaine | 0 (no) | 0 (no) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Barker et al., 2017 | Contingency degradation | 0 (no) | 0 (no) | 1 (lever press, licking central port) | 1 (unatural) | 0 (no) | 0 (no) | 0 (no) | 1 | 3 |
| Beier et al., Cell, 2015 | intracranial-self stimulation of mPFC->VTA projections | 0 (no) | 0 (no) | 5 ports | 0 (nose poking) | 0 (no) | 0 (no) | 0 (no) | 1 | 6 |
| Berg et al., 2017 | Open field / Elevated plus maze | 0 (no) | 0 (no) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Bolkan et al., Nat Neurosci, 2017 | DNMTS T maze | 0 (no) | 0 (no) | 2 (go right or left) | 0 (normal behavior) | 0 (no) | 1(20-60s) | 0 (no) | 1 | 4 |
| Breton-Provencher and Sur, Nat Neurosci, 2019 | Pupil dilatation | 1 (Sound) | 0 (pure tone) | 0 (nothing to do) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 0 | 1 |
| Bukalo et al., Neuropsychology, 2015 | Contextual Fear conditioning | 2(fear cage, footshock) | 1 (full context) | 1 (freezing) | 0 (normal aversive behavior) | 0 (no) | 0 (no) | 1 (1day) | 1 | 5 |
| Carus-Cadavieco et al., 2017 | Food rewarded learning task | 0 (no) | 0 (no) | 1 (nose poke, eating, drinking) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Cho et al., BioRxiv, 2019 | attention set shifting task | 2 (olfactory and somatosensory) | 0 natural cues | 1 (dig reward) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 4 |
| Cho et al., Neuron, 2015 | attention set shifting task | 2 (olfactory and somatosensory) | 0 natural cues | 1 (dig reward) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 4 |
| Courtin et al., Nature, 2014 | Fear conditioning | 2 (sound, CS, footshock, US) | 0 (pure tone) | 1 (freezing) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (24h) | 1 | 6 |
| Dejean et al., 2016 | Fear conditioning | 2 (sound, CS, footshock, US) | 0 (pure tone) | 1 (freezing) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (24h) | 1 | 5 |
| Esmaeili et al., 2020 | delayed whisker detection task | 3 (visual,whisker,aud)) | 0 (no) | 1 (licking) | 0 (normal behavior) | 0 (no) | 1 (1s) | 0 (no) | 0 | 5 |
| Felix-Ortiz et al., Neuroscience, 2016 | social interaction | 1 juvenile mouse | 2(complex) | 1(social) | 1(Social behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 6 |
| Gilad et al., Neuron, 2018 | Texture discrimination | 2 (textures) | 1 (more natural stimulus) | 1(lick) | 0 (normal behavior) | 1 (Sound) | 1(1,2-4,5s) | 0 (no) | 0 | 6 |
| Goard et al., Elife, 2016 | Go/NoGo delayed visual task | 2 (visual gratings) | 1 (gratings) | 1(lick) | 0 (normal behavior) | 1 (sound) | 1 (0,3,6) | 0 (no) | 0 | 6 |
| Granon and Changeux, Behav Brain Res, 2012 | Spatial learning and novel object and change of the baited arm (flexibility) | 0 (no) | 0 (no) | 2 (go right or left) | 0 (normal behavior) | 0 (no) | 0 (no) | 1(2min) | 1 | 4 |
| Guo et al., Neuron, 2014 | whisker-based object localization task with a delayed, directional licking response | 2 (pole positions) | 1 (object localisation with whisker close or far) | 2 (lick rigth, lick left) | 0 (normal behavior) | 1 (Sound) | 1 (3s) | 0 (no) | 0 | 7 |
| Gutzeit et al., 2020 | Contextual Fear conditioning | 2(fear cage, footshock) | 1 (full context) | 1 (freezing) | 0 (normal aversive behavior) | 0 (no) | 0 (no) | 1 (1day) | 1 | 5 |
| Halladay et al., 2020 | Footshock punished EtOh self administration | 1 (footshock, US) | 0 (no) | 2 (lever press, licking central port) | 1 (unatural) | 0 (no) | 0 (no) | 0 (no) | 1 | 5 |
| Hare et al., 2019 | Novelty supressed feeding | 0 (no) | 0 (no) | 1 (eating) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Heikenfeld et al., 2020 | Spatial non match to sample | 0 (no) | 0 (no) | 2 (go right or left) | 0 (normal behavior) | 0 (no) | 1 (20s) | 0 (no) | 1 | 4 |
| Hu et al., Neuron, 2019 | Go/No Go Visual discrimination task | 2 (visual gratings) | 1 (gratings) | 1(lick) | 0 (normal behavior) | 1 (Sound) | 0 (no) | 0 (no) | 0 | 5 |
| Huang et al., 2019 | Paw withdrawal | 1 (nociceptive, paw) | 0 (no) | 1 (paw withdrawal) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 3 |
| Huda et al., BioRxiv, 2018 | Visual forepaw orienting task | 2 (left - right) | 1 (square on a screen) | 2 (left right) | 1 (moving a trackball) | 1(sound) | 1 (500ms) | 0 (no) | 0 | 8 |
| Inagaki et al., J.Neurosci, 2018 | Auditory delayed discrimination task | 2(sounds) | 0 (pure tone) | 2 (lick right or left) | 0 (normal behavior) | 1 (Sound) | 1(1,2s) | 0 (no) | 0 | 6 |
| Inagaki et al., Nature, 2019 | Auditory delayed discrimination task | 2(sounds) | 0 (pure tone) | 2 (lick right or left) | 0 (normal behavior) | 1 (Sound) | 1(1,2s) | 0 (no) | 0 | 6 |
| Itokazu et al., Nat Commun, 2018 | Sacade task in Mice ! | 2 LEDs | 0 (pure) | 2 (saccade forward or backward) | 1 (unatural inhead fixed) | 1 (fixation LED) | 0 (no) | 0 (no) | 0 | 6 |
| Jennings et al., Nature, 2019 | Apetitive reward or social stimulus with lick behaviro | 1 | 2(social) | 1(lick) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 0 | 4 |
| Jhang et al., 2018 | Fox urine avoidance | 1 (olfactory) | 0 (natural odor) | 1 (freezing) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 3 |
| Kamigaki and Dan, Nat Neurosci, 2017 | Go/NoGo delayed auditory task and 2AFC delayed | 1 (auditory) | 0 (pure tone) | 2 (lick and left/right) | 0 (normal behavior) | 0 (no) | 1 (5s) | 0 (no) | 0 | 4 |
| Kim et al., 2016 | Fear conditioning | 2 (sound, CS, footshock, US) | 0 (no) | 1 (freezing) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (2days-4days) | 1 | 5 |
| Kim et al., Cell, 2016a | Spatial Woring memory : delayed non-match-to-place task. | 2 (left - right) | 0 (visit arm) | 2 (left right) | 0 (visit arm) | 0 (door opening ?) | 1 (3s) | 1 (previous trial) | 1 | 8 |
| Kim et al., Cell, 2016b | 3CSRTT | 3 (visual) | 0 (flashing LED) | 3 (Right,Center,Left) | 0 (nose poking) | 0 but trial initiation so 1 | 1 (3-5s) | 0 (no) | 1 | 8 |
| Kim et al., Cell, 2017 | Lever-press reward seeking suppression | 1 (auditory) | 0 (pure tone) | 1 (press or not) | 1(lever pressing) | 0 (no) | 0 (no) | 1 (yes sometimes there is a shock) | 0 | 4 |
| Kingsbury et al., 2020 | Social exploration assay | 2 (novel male and female) | 1 (complex) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 5 |
| Klavir et al., Nat Neurosci, 2017 | Fear conditioning | 2 (sound, CS, footshock, US) | 0 (pure tone) | 1 (freezing) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (24h) | 1 | 5 |
| Koike et al., Neuropsychopharmacology, 2016 | 5CSRTT | 5 (touch-screen squares) | 1 (pure ligth) | 5 (nose poke locations) | 0 (normal behavior) | 1 (house light at trial onset) | 0 (no) | 0 (no) | 1 | 10 |
| Kumar et al., 2013 | elevated plus maze | 0 (no) | 0 (no) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Lak et al., BioRxiv, 2018 | 2-alternative visual decision making task with switching reward contigency | 2 (visual gratings) | 2 (gratings and intensity modulation) | 2 (turn steerwheel rigth or left) | 1 (unatural) | 0 (no) | 0 (no) | 0 (no) | 0 | 7 |
| Le Merre et al., Neuron, 2018 | Whisker-based detection task | 1 (whisker deflection) | 0 (1ms deflection) | 1 (licking) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 0 | 2 |
| Lee et al., Neuron, 2019 | elevated plus maze | 0 (no) | 0 (no) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Leinweber et al., Neuron, 2017 | 2D Virtual navigation task and reversal of the optic folw (mismatch) | 1 virtual reality tunnel | 2 (circles on the walls and reversion of visual flow) | 2 running left rigth on a track ball | 1 (reversion of the visual flow) | 0 (no) | 0 (no) | 0 (no) | 0 | 6 |
| Li et al., 2020 | Delayed Go/noGo Auditory | 3 (target, non target tone, airpuff/electrical shock form the lick spout) | 0 (pure tone) | 1 (licking) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (0-2s) | 0 | 5 |
| Li et al., Nature, 2015 | whisker-based object localization task with a delayed, directional licking response | 2 (pole positions) | 1 (object localisation with whisker close or far) | 2 (lick rigth, lick left) | 0 (normal behavior) | 1 (Sound) | 1 (3s) | 0 (no) | 0 | 7 |
| Li et al., Nature, 2016 | whisker-based object localization task with a delayed, directional licking response | 2 (pole positions) | 1 (object localisation with whisker close or far) | 2 (lick rigth, lick left) | 0 (normal behavior) | 1 (Sound) | 1 (3s) | 0 (no) | 0 | 7 |
| Liu et al., 2016 | Sucrose self administration | 1 (light cs) | 0 (flashing LED) | 1 (lever press) | 1 (unatural) | 0 (no) | 0 (no) | 0 (no) | 1 | 4 |
| Liu et al., Science, 2014 | Olfactory delayed nonmatch to sample. Olfactory non-delayed nonmatch to sample and Go/noGo olfactory discrimination | 4 (2 odors, 4 combinations) | 0 (pure odor) | 1 (licking) | 0 (normal behavior) | 0 (no) | 1 (4-5s) | 0 (no) | 0 | 6 |
| Makino et al., Neuron, 2017 | Lever-press (auditory cued) motor task | 1 (auditory) | 0 (pure tone) | 2 (lever pressing and licking) | 1 (unatural) | 0 (no) | 0 (no) | 0 (no) | 0 | 4 |
| Manita et al., Neuron, 2015 | Spontaneous Test Preference | 2 (textures) | 1 (more natural stimulus) | 0 (nothing to do) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 3 |
| Manita et al., Neuron, 2015 | Tactile discrimination task | 2 (textures) | 1 (more natural stimulus) | 2 (go right or left) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 5 |
| Marton et al., J. of Neuro, 2018 | attention shifting task | 2 (sound and ligth) | 0 (pure tone) | 1 (dig reward) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 4 |
| Mayrhofer et al., Neuron, 2191 | perceptual decision making | 2 (whisker and Auditory) | 0 Natural deflection and pure tone) | 1 (licking) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 0 | 3 |
| Morandel et al., Scientific reports, 2017 | directional joystick task | 2 (vibrations) | 1 (more natural stimulus) | 2 (pull or push joystick) | 1(unatural) | 2 (Go and reward sounds) | 1(1s) | 0 (no) | 0 | 9 |
| Morawska and Fendt, 2012 | Fear conditioning | 2 (sound, CS, footshock, US) | 0 (pure tone) | 1 (freezing) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (24h) | 1 | 5 |
| Murugan et al., Cell, 2017 | exploration of 3-chamber arena with a social target vs object | 3 (complex : novel object or other mouse) | 1 (other mouse, natural social behavior) | 1 (moving around) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (1 day) | 1 | 6 |
| Nakajima et al., 2019 | 2AFC (auditory vs visual) with rule instruction before | 3 (one visual and 2 auditory) | 1 (LED and upsweep/downsweep) | 2 (go right or left) | 1 (Rule instructed) | 2 (high and low pitch noise) | 1 (500-700ms) | 0 (no) | 1 | 10 |
| Nakayama et al., 2018 | Probabilistic reversal task | 1 (light) | 0 (flashing LED) | 2 (go right or left) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 4 |
| Namboodiri et al., 2019 | Reward conditioning | 2 (auditory) | 0 (pure tone) | 1 (licking) | 0 (normal behavior) | 0 (no) | 1 (3s but they can lick) | 0 (no) | 0 | 4 |
| Newmyer et al., 2019 | Food intake assay | 0 (no) | 0 (no) | 1 (feeding) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Nguyen et al., 2020 | Olfactory delayed nonmatch to sample | 2 Odors | 0 natural cues | 1 (dig reward) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (5s) | 1 | 5 |
| Otis et al., Nature, 2017 | Reward conditioning | 2 (auditory) | 0 (pure tone) | 1 (licking) | 0 (normal behavior) | 0 (no) | 1 (3s but they can lick) | 0 (no) | 0 | 4 |
| Padilla-Coreano et al., Neuron, 2019 | elevated plus maze | 0 (no) | 0 (no) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Pascoli et al., Nature, 2018 | Self stimulation of the VTA and perseverance when foot shock are given | 0 (no) | 0 (no) | 2 (levers) | 1 (unatural) | 0 (no) | 0 (no) | 1 (24h) | 1 | 5 |
| Pinto and Dan, Neuron, 2015 | Auditory discrimination task | 2 (auditory) | 0 (pure tone) | 1 (licking) | 0 (normal behavior) | 1 (visual) | 1 (1s) | 0 (no) | 0 | 5 |
| Pinto et al., Cell, 2019 | visual accumulation task | 1 visual tower | 3 (Virtual reality) | 2(running and lick) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 0 | 6 |
| Pinto et al., Cell, 2019 | visual memory guided task | 1 visual cue | 3 (Virtual reality) | 2(running and lick) | 0 (normal behavior) | 0 (no) | 1 | 0 (no) | 0 | 7 |
| Popescu et al., PNAS, 2016 | DA-paired stimulus (pavlovian) | 1 (auditory) | 0 (pure tone) | 1 (licking) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 0 | 2 |
| Rajasethupathy et al., 2015 | Contextual fear conditioning | 2(fear cage, footshock) | 1 (full context) | 1 (freezing) | 0 (normal aversive behavior) | 0 (no) | 0 (no) | 1 (1day) | 1 | 6 |
| Rikhye et al., Nat Neurosci, 2018 | Attentional cued switching task | 4 (2noise,2visual) | 1(sweeps or pure LED) | 2 (go right or left) | 0 (normal behavior) | 2 (Sounds defining contexts) | 1(900ms) | 0 (no) | 1 | 10 |
| Rozeske et al., Neuron, 2018 | Fear condiriotioning with different contexts (Auditory, visual, olfactory) | 3 (context defined by white noise, lime odor and house light) | 1 (context) | 1 (freezing) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (24h) | 1 | 7 |
| Schmitt et al., Nature, 2017 | 2AFC (auditory vs visual) with rule instruction before | 3 (one visual and 2 auditory) | 1 (LED and upsweep/downsweep) | 2 (go right or left) | 1 (Rule instructed) | 2 (high and low pitch noise) | 1 (400ms) | 0 (no) | 1 | 10 |
| Schneider et al., Nature, 2014 | Running on a treadmill | 0 (no) | 0 (no) | 1 (running) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Sheggia et al., 2020 | Affective state discrimination task | 1 (other mice stressed or water restricted) | 2 (complex social) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 5 |
| Siniscalchi et al, Nat Neurosci, 2016 | Adaptative decision making task (=rule switching) | 2 (auditory) | 1 (upsweep/downsweep) | 2 (lick right or left) | 1 (Rule instructed) | 1 (sound) | 0 (no) | 0 (no) | 0 | 7 |
| Spellman et al., BioRxiv, 2019 | Serial Extradimensional shifting | 4 (2 whisker, 2 odors) | 0 (no) | 2 (lick right or left) | 0 (normal behavior) | 1 (white noise) | 0 (no) | 0 (no) | 0 | 7 |
| Spellman et al., Nature, 2015 | Spatial Delayed non-match to place | 0 (no) | 0 (no) | 2 (go right or left) | 0 (normal behavior) | 0 (no) | 1(10-60s) | 0 (no) | 1 | 4 |
| Starkweather et al., Neuron, 2018 | Olfactory pavlovian condition with 100% and 90% reward probability | 1 (odor) | 0 (pure odor) | 1 (licking) | 0 (normal behavior) | 0 (no) | 1 (1.2-2.8s) | 0 (no) | 0 | 3 |
| Tamura et al., Nat Commun, 2017 | Spatial Delayed non-match to place | 0 (no) | 0 (no) | 2 (go right or left) | 0 (normal behavior) | 0 (no) | 1(10-60s) | 0 (no) | 1 | 4 |
| Tan et al., 2019 | Social novelty test | 2 (other mice novel familiar) | 2 (complex social) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 6 |
| Tuscher et al., 2018 | Object placement (or recognition) | 2 (objects) | 0 natural cues | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (4h - 24h) | 1 | 5 |
| Van den Oever et al., 2013 | Conditioned place preference | 2 (spatial compartements) | 0 (visual cues) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (yes) | 1 | 5 |
| Vertechi et al., 2020 | probabilistic foraging | 0 (no) | 0 (no) | 2 (exploration, nose poke) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 3 |
| White et al., Cell Rep, 2018 | 5CSRTT | 5 (touch-screen squares) | 1 (pure ligth) | 5 (nose poke locations) | 0 (normal behavior) | 1 (house light at trial onset) | 0 (no) | 0 (no) | 1 | 10 |
| Wimmer et al., Nature, 2015 | 2AFC (auditory vs visual) with rule instruction before | 3 (one visual and 2 auditory) | 1 (LED and upsweep/downsweep) | 2 (go right or left) | 1 (Rule instructed) | 2 (high and low pitch noise) | 1 (500-700ms) | 0 (no) | 1 | 10 |
| Wu et al., 2020 | DNMTS | 4 (2 odors, 4 combinations) | 0 (pure odor) | 2 (lick and left/right) | 0 (normal behavior) | 1(go cue, pure tone) | 1(1-4s) | 0 (no) | 0 | 8 |
| Xu et al., BioRxiv 2019 | Sequence licking task | 1 (Sound) | 0 (no) | 1 (licking) | 1(sequence) | 0 (no) | 0 (no) | 0 (no) | 0 | 3 |
| Yamamuro et al., 2020 | Novel object/mouse test | 2 (object-mouse) | 2 (complex social) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 6 |
| Ye et al., Cell, 2016 | Place preference | 0 (no) | 0 (no) | 1 (running) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 2 |
| Yizhar et al., Nature, 2011 | exploration of 3-chamber arena with a social target vs object | 3 (complex : novel object or other mouse) | 1 (other mouse, natural social behavior) | 1 (moving around) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 5 |
| Zhang et al., 2015 | Conditioned place preference | 2 (spatial compartements) | 0 (visual cues) | 1 (exploration) | 0 (normal behavior) | 0 (no) | 0 (no) | 0 (no) | 1 | 4 |
| Zhang et al., Science, 2014 | Visual discriminaiton task | 2 (visual gratings) | 1 (gratings) | 1(lick) | 0 (normal behavior) | 1 (sound) | 0 (no) | 0 (no) | 0 | 5 |
| Zhu et al., 2020 | Delayed olfactory paired association task | 4 (2 odors, 4 combinations) | 0 (pure odor) | 1 (licking) | 0 (normal behavior) | 0 (no) | 1 (5-10s) | 0 (no) | 0 | 6 |
| Zimmermann et al., Biol. Psychiatry, 2017 | Reinforcement learning | 0 (no) | 0 (no) | 2 (go right or left) | 0 (normal behavior) | 0 (no) | 0 (no) | 1 (24h) | 1 | 4 |
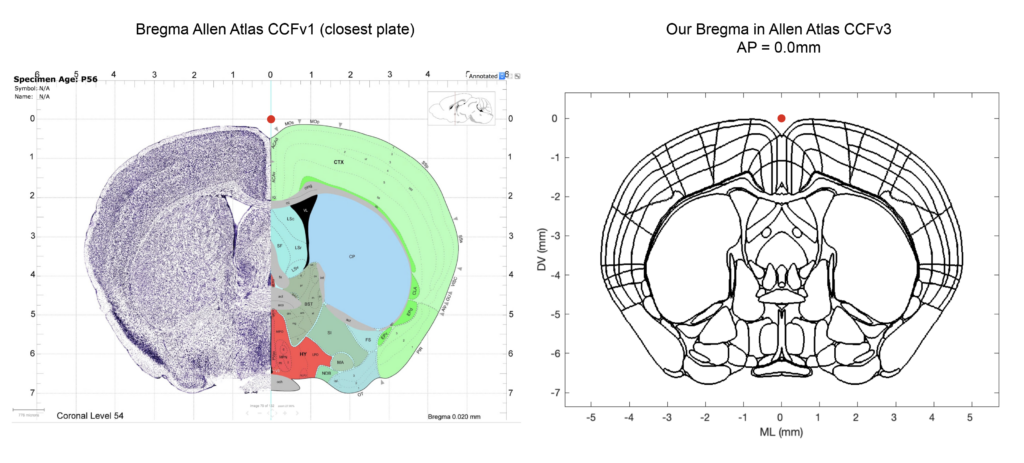
Bregma in CCFv3
The Allen Institute explicitly state that they do not have a definition for Bregma for the ~1600 adult mouse brains used to construct the CCFv3: “Each one of those brains was dissected from a skull, and every skull has a slightly different Bregma location; hence the CCF does not have a single brain-in-skull as a source (which would have a corresponding Bregma coordinate), but is made from brains out-of-skull that have been registered to achieve an ‘average’ volume that is defined by the brain itself, ex cranio. “ (source here). Therefore we had to place Bregma ourselves in the CCFv3. We placed Bregma based on the information available in the first version (CCFv1) of the ARA and how others labs have placed it (see the great Github repo of the Cortex lab from where we derived our scripts). Below, you can appreciate graphically where we place Bregma in the CCFv3. The matlab script “AllenCCFBregma.m” is carefully commented in regards to our coordinate triplet to provide transparency and reproducibility for other users.
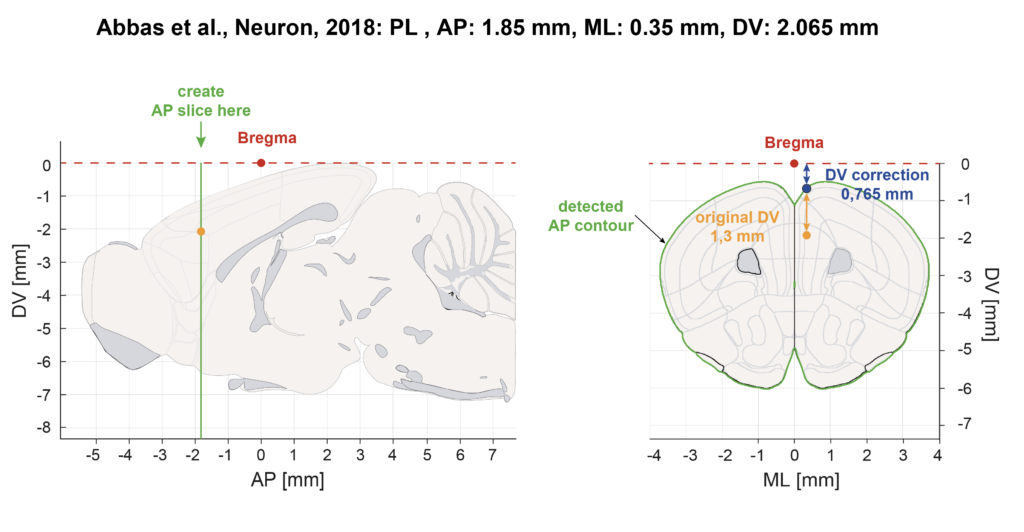
DV Correction
Experimenters report the Bregma DV coordinate either relative to Brain surface or relative to Bregma 0 (skull landmark). In the Table1-Database we performed DV corrections to obtain the DV coordinate into the CCFv3. When Bregma was reported from the brain surface, we performed a correction with the matlab script “Bregma_DV_correction.mat”. This script plots the contours of the coronal section for the AP and ML coordinates provided in the referenced publication and finds the distance between brain the brain surface and the CCFv3 Bregma. This value needs to be added to the mean DV value provided in the original publication: column ”Brain surface DV Corr” in Table1-Database. When Bregma was reported from Bregma 0 (skull landmark) no correction is needed.
Limitations
The CCFv3 version of the Allen Atlas is a 10um voxel resolution space and our analysis using this Atlas has some limitations. We generated 3D meshes for the brain outline, for each PFC region (MOs, ACA, PL, ILA, ORBm, ORBvl and ORBl) and for the PFC sudivisions described in the section of our Perspective ”Parcellations of the PFC” (dmPFC, vmPFC and vlPFC) by using the MATLAB ”isosurface.m” function (see folder Brain_meshes). To detect in which Allen PFC region each publication was peformed we used the ”inpolyhedron.m” function (Copyright © 2015, Sven Holcombe). The thickness of the borders in the actual version of the CCFv3 is around 30um (3 voxels). As a result some triplet of coordinates are allocated to 2 regions while performing our “inpolyhedron.m” detection procedure. We inspected manually each triplet of coordinates whithin its respective ARA plate (see folder AllenAtlasLocationCCFv3_individualstudies) and visually assesed for these ambiguous cases to which Allen PFC region they belong to. Our level of incertitude regarding PFC region assignment needs to be put into perspective because, as accurate as we can be as experimenters, the provided stereotaxic coordinates in every publication are only averaged values reflecting the mean location of the targetted brain structures. This experimental incertitude is by itself an important limitation of our analysis and needs to be taken into consideration.
Source
© 2015 Allen Institute for Brain Science. Allen Mouse Brain Atlas (2015) with region annotations (2017).
A whole brain atlas of the monosynaptic input targeting four different cell-types in the medial prefrontal cortex of the mouse
The DMC Drive
Developed by Hoseok Kim, PhD, CarlenLab, Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden, Aug. 2019
General description
The DMC-drive is a small, light and simple positioning system for chronic electrophysiology experiments using multiple tetrodes in conjunction with optogenetics in mice and rats. The drive is composed of three parts that can be manufactured easily and at low cost using a CAD design file (Autodesk Inventor) and a 3D printer.
Download the files here.
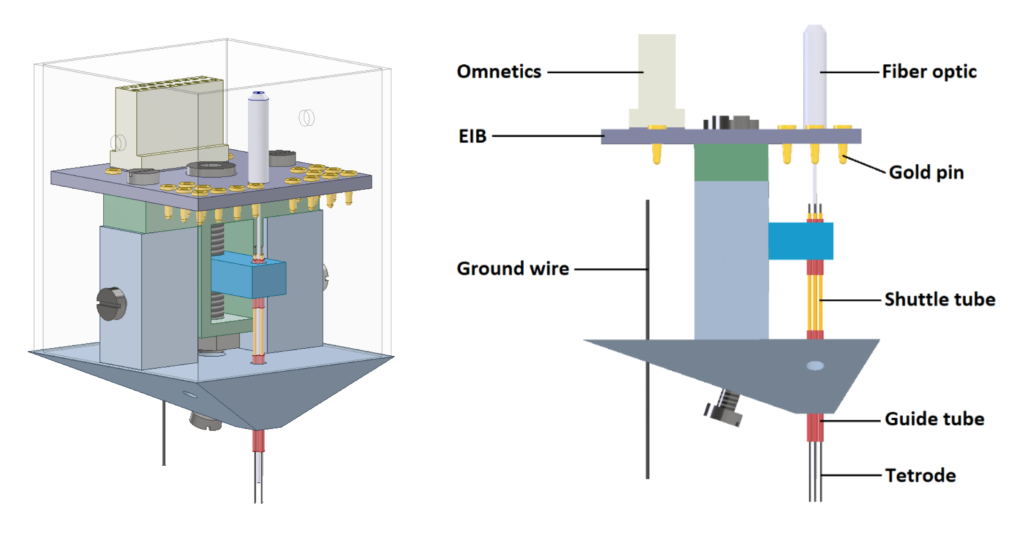
Specifications
- Model name: DMC-drive
- Weight (without EIB): < 2g
- Size (L x W x H): 16 x 16 x 22mm
- Max. number of electrodes: 16 (4 tetrodes)
- Max. electrode travel distance: 4mm
- Screws & nut: M1.6-10mm hex socket cap head x1 (shuttle screw) / M1.6-thin hex nut x1 (shuttle nut) / M1-5mm slotted cheese head x2 (fixing screw for EIB, shuttle, and body part) / M1-2mm slotted cheese head x3 (fixing/anchor screw)
- Species: mouse or rat
Parts
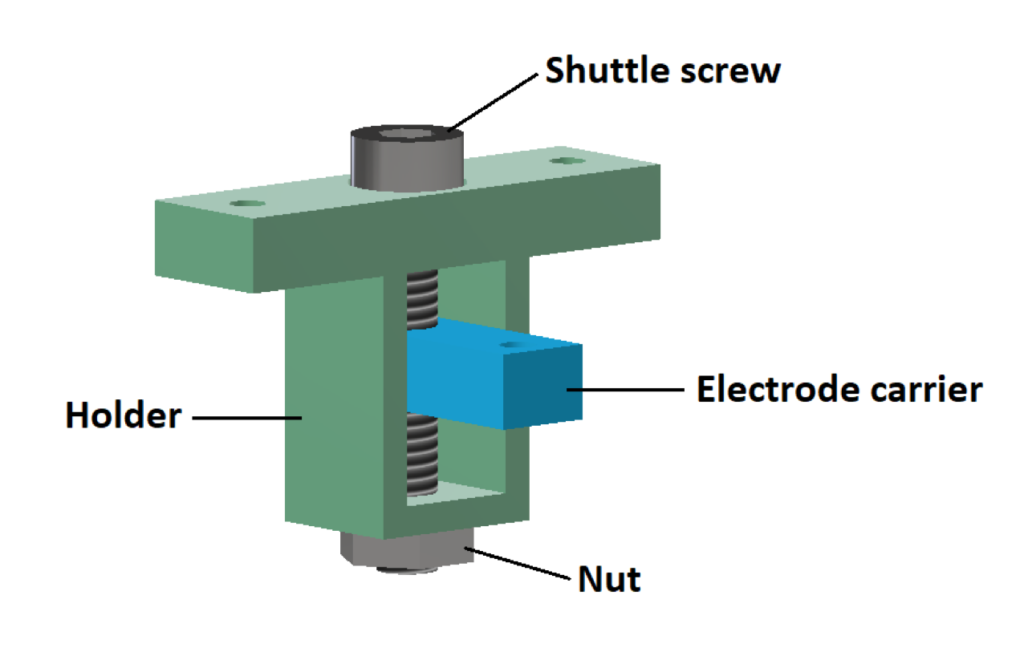
- Shuttle (electrode carrier, holder)
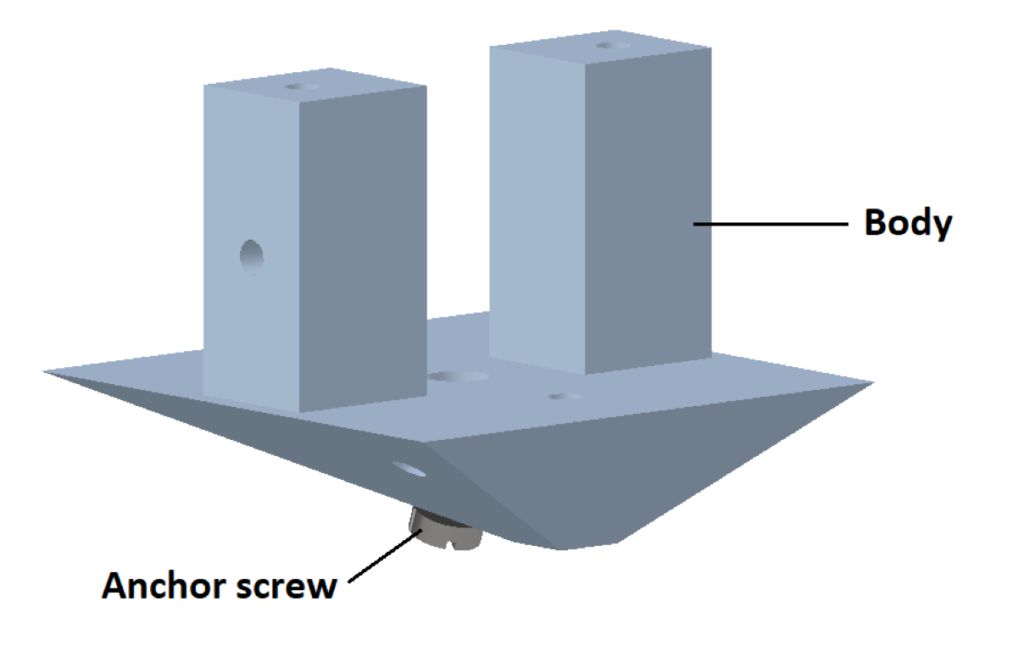
2. Body
3. Case